Who are the users of BRCA Exchange, what features do they value, and what are their needs? To assess these questions, we ran on online survey of the BRCA Exchange users, from Jaunary 22, 2023 to August 22. During this time, 160 individuals responded to the survey. While this is a small fraction of the overall BRCA Exchange usage, as measured by Google Analytics, it’s also large enough to arguably offer a representative sample. This article describes what we learned from this survey.
Who were the survey respondents?
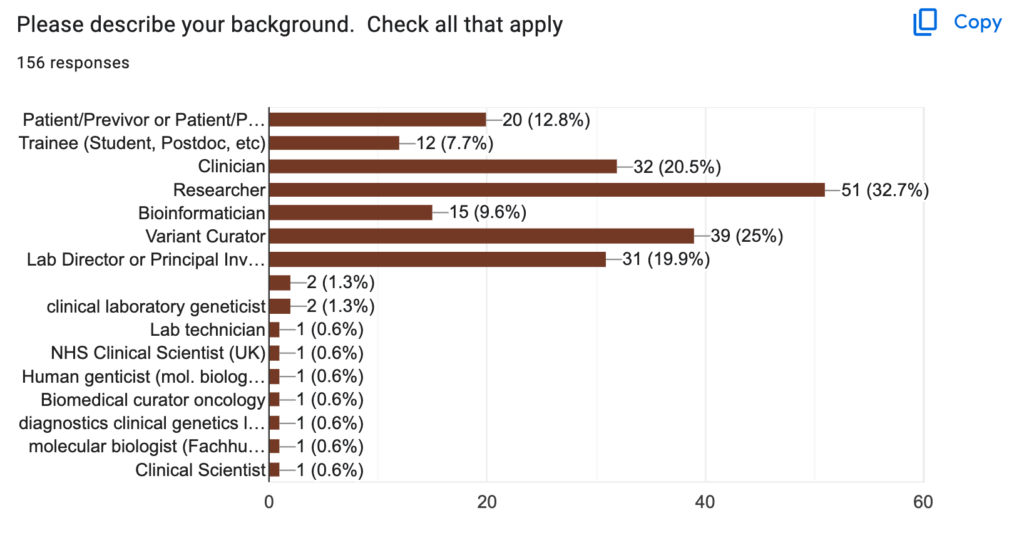
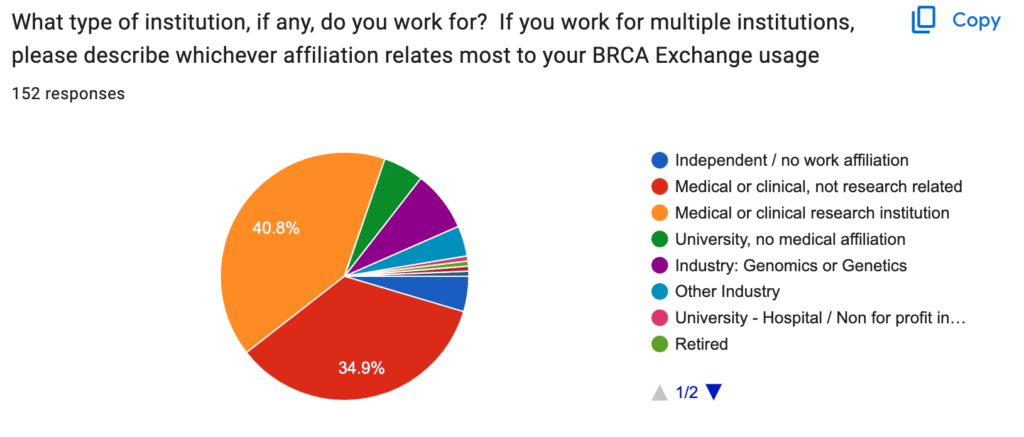
Most respondents were professionals involved in variant assessment at medical or clinical institutions, with a balance of research institutions and non-research institutions. They were mostly researchers, variant curators, clinicians and lab directors. Yet a surprising proportion (12.6%) of the respondents were patients or previvors, despite the highly technical content of the site.
Most respondents were frequent users, who visit the site once a month or more. Yet 19% of the respondents were first-time users, and 13% were infrequent users (who visit the site less than once per month). These first-time and infrequent users included many patients/previvors and many trainees.
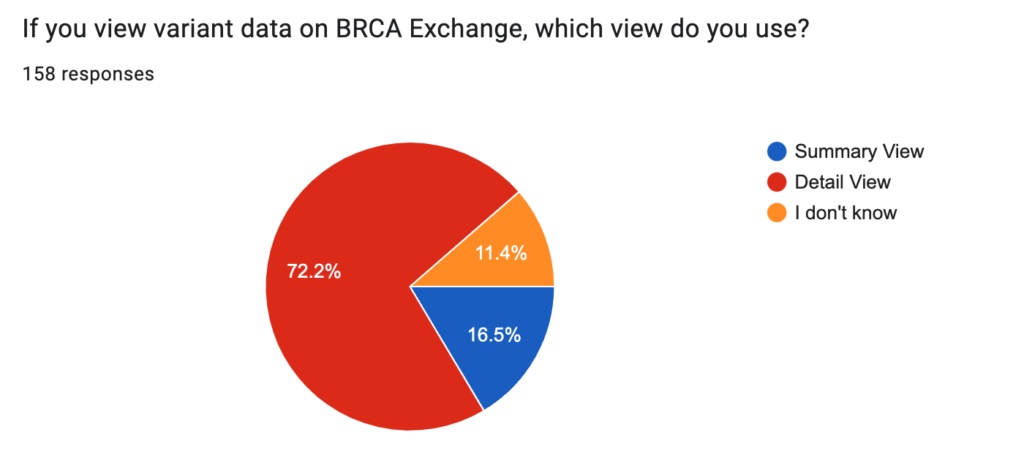
BRCA Exchange contains two views, or display modes. The default view is the Summary view, which contains only the ENIGMA expert panel classification and the variant nomenclature tile (i.e. it identifies the variant and presents the expert panel classification, if any). Users who select to see more data can toggle to the Detail view, which presents additional data including clinical classifications from ClinVar and LOVD (some of which can be contradictory), and variant curation resources including literature search results, allele frequencies and curated functional assay results. Most respondents indicated that they use the Detail view. First-time users often indicated that they didn’t know what view they were using, and were undoubtedly using the Summary view. Surprisingly, many of the respondents who indicated that they use the Summary view also indicated that they visit the site several times per week. This suggests that there is a demographic that values getting only the most important variant details, and no more.
We asked those users who use the Detail view to indicate which features they found most valuable, ranking them from Not Valuable to Essential. Surprisingly, the top-ranked features were the ENIGMA and ClinVar variant curations: surprising because these featues are available in many places, including ClinVar. Yet it’s been pointed out that maybe this result is not so surprising, because ClinVar by its nature has to be a one-size-fits-all resource, while BRCA Exchange targets a specific user group and can have a more tailored user interface.
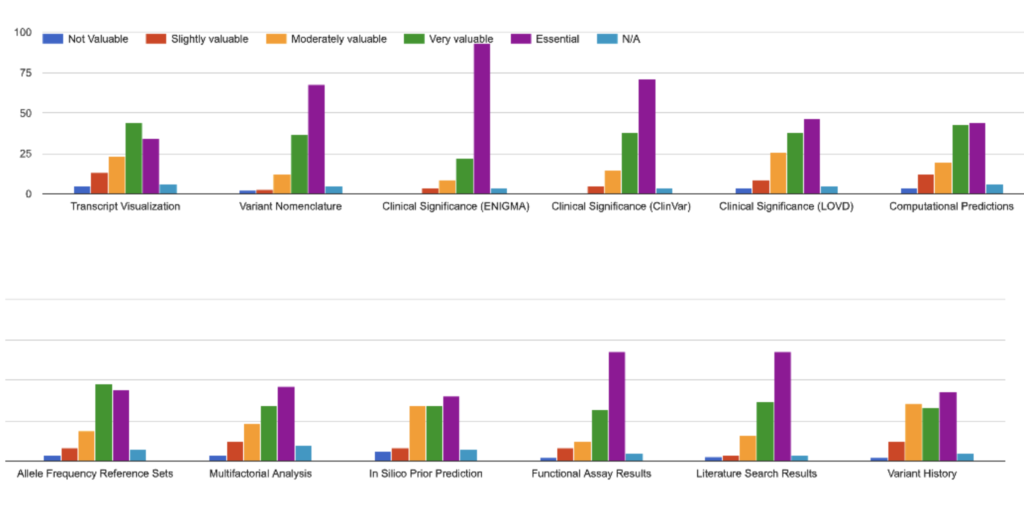
Other highly-valued features included the Literature Search Results and Functional Assay Results, data which are not readily available via other resources. For contrast, the widely-available Allele Frequency Reference Data (from gnomAD), Computational Predictions (which include BayesDel and SpliceAI) and in Silico Prior Prediction tiles were rated as less valuable overall: these tiles contain data which are available from many sources, and which tend to be static (i.e. if a user downloads or records them once, there’s no need to review the tile again).
We asked the users if there are any other features they would like which are not currently available on the site. This question received 43 responses. The feature requested most frequently was the proposed ACMG/AMP evidence codes; this was requested by eight respondents. These codes signify when a VUS has significant evidence in some category to be reclassified, as described by the ACMG/AMP variant curation standards and the ClinGen BRCA Variant Curation Expert Panel (VCEP). For each distinct form of evidence (functional assay results, population frequencies, etc), one or more evidence codes indicates when the evidence appears significant by the VCEP standards and can contribute to VUS reclassification. While ClinGen stipulates that these evidence codes should be formally assigned and reviewed by biocurators, the provisional evidence codes (those awaiting biocuration) can still provide valuable information for clinicians and researchers who are evaluating BRCA variants. Other new feature requests include new help material for non-expert users, greater VUS classification, resources to help users find further information on their own variants, and links to external resources including HGMD and dbSNP.
Finally, we asked the users if there was anything else they would like to share. 32 users responded. Here were some of these responses.
To close, we wish to thank all of the respondents for their time and thank them for this invaluable feedback, which is already helping us prioritize the development of new features!